mRNA, lncRNA and circRNA in human biofluids
Welcome to exoRBase 3.0
exoRBase is a repository of messenger RNA (mRNA), long non-coding RNA (lncRNA) and circular RNA (circRNA) derived from RNA-seq data analyses of human biofluids. Since its pioneering launch as exoRBase 1.0 (2017) with exosomal RNAs from blood, the database has become indispensable for liquid biopsy research. exoRBase 2.0 (2021) expanded significantly to 905 exLR-seq datasets across four biofluids, paving the way for broader comparative studies.
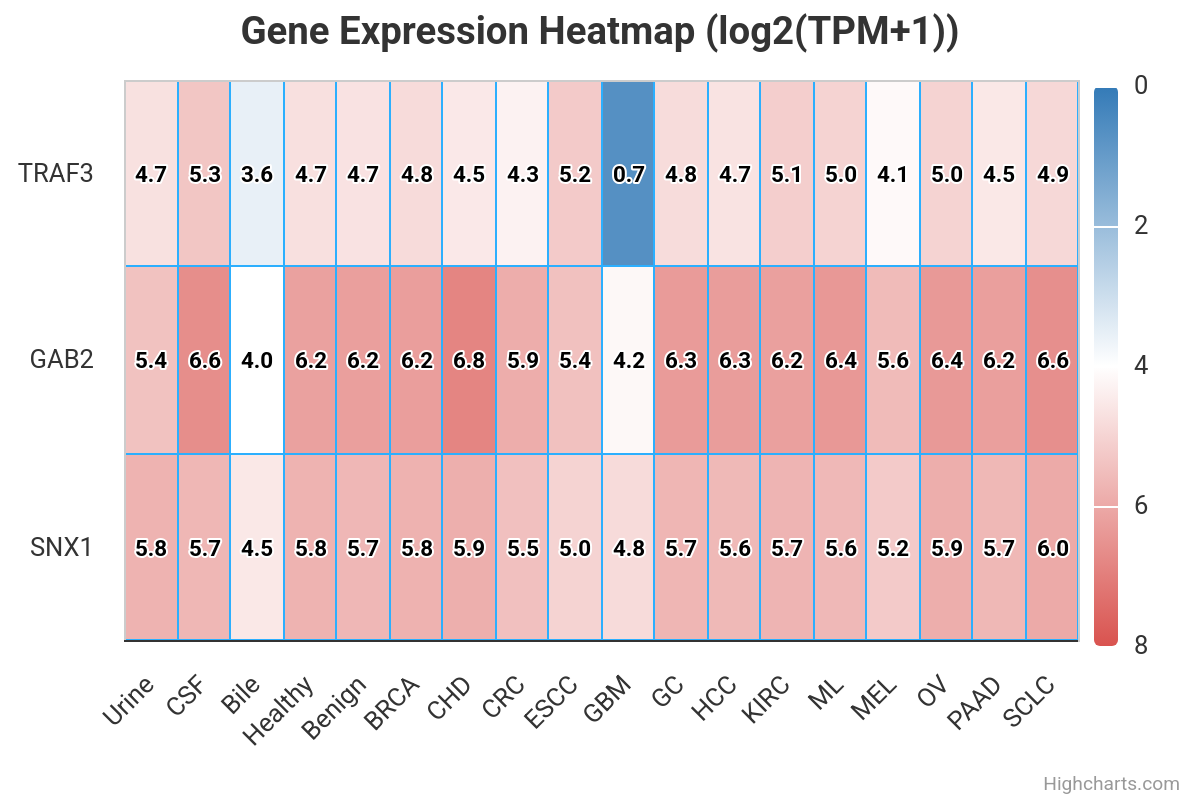
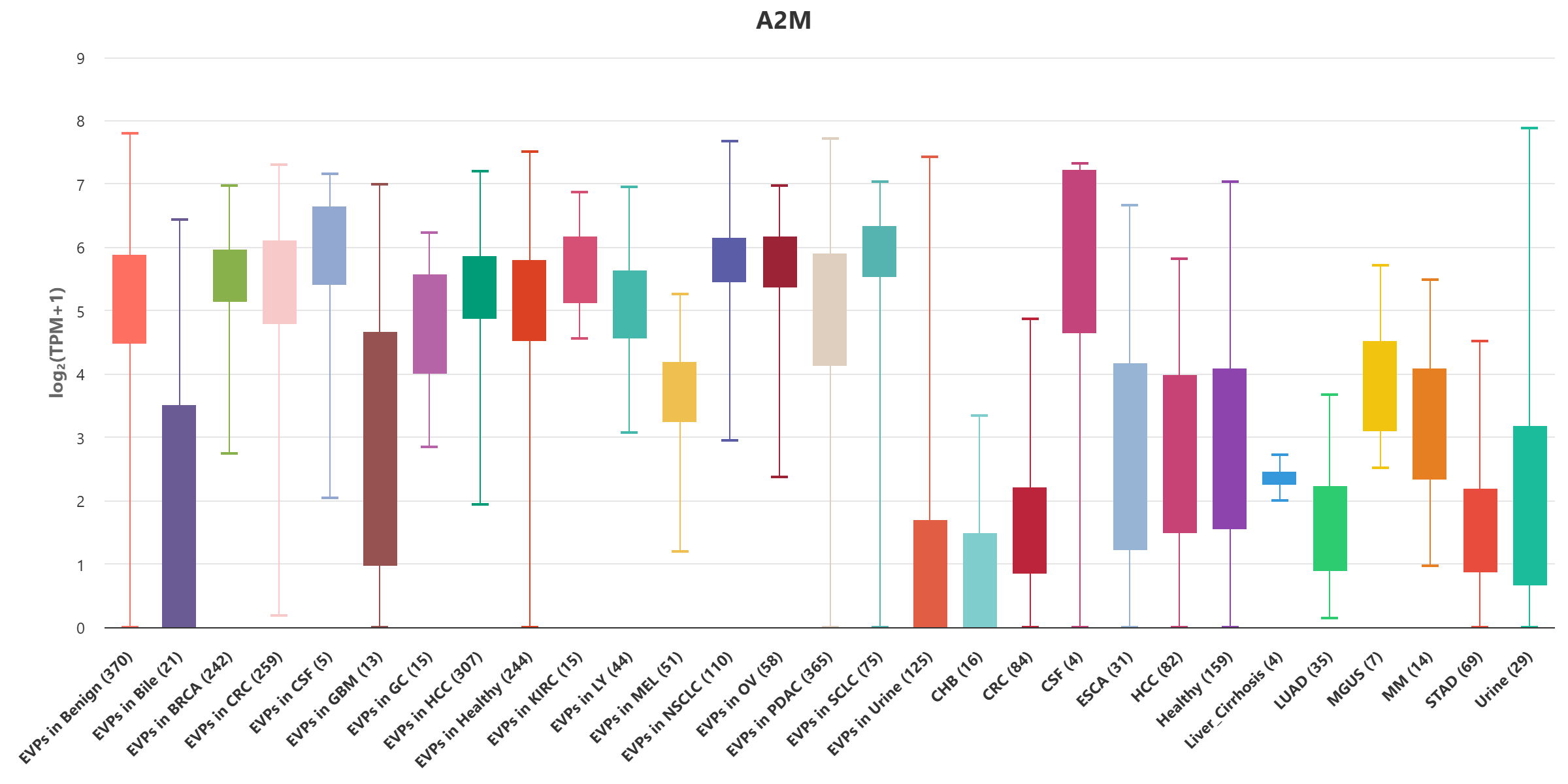
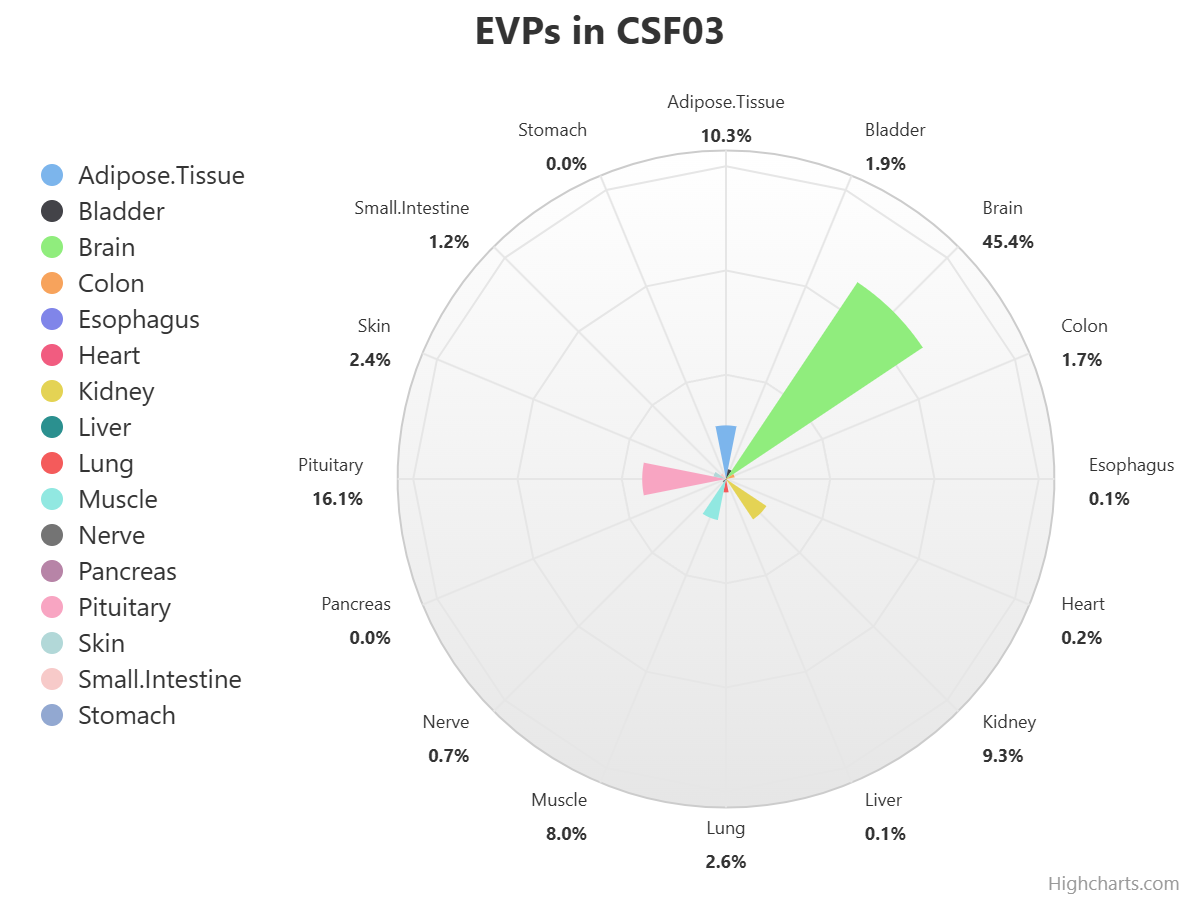
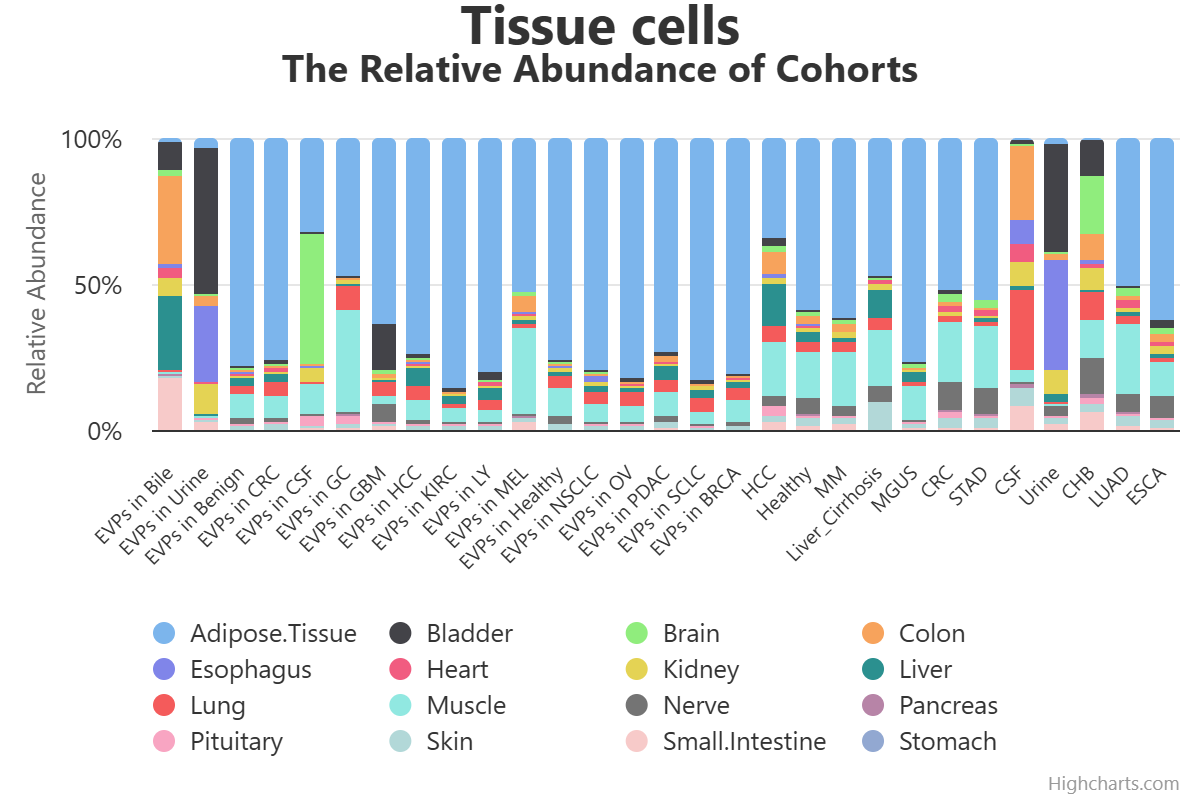
Here, we present exoRBase 3.0 – a transformative update integrating approximately 3,000 samples spanning Extracellular Vesicles and Particles (EVPs) and cell-free RNAs (cfRNAs) across diverse human biofluids. This major expansion unifies EVP-associated and cell-free long RNAs under the extracellular long RNA (exLR) framework. exoRBase 3.0 features the integration and visualization of RNA expression profiles, as well as the functional pathway-level changes and the heterogeneity of circulating RNA origins, reflecting critical advances in extracellular RNA biology and their established utility in biomarker discovery for precision medicine.
Release Notes&News
- exoRBase 3.0 is published in Database Issue of Nucleic Acids Research. [October 2025] [Full Text]
- exoRBase has been updated to verson 3.0. [June 2025] [Version 2.0]
- 594 cfRNA-seq samples from 7 experiments have been integrated into exoRBase. [June 2025]
- Circular RNA landscape in extracellular vesicles from human biofluids, published in Genome Medicine. [October 2024] [Full Text]
- exoRBase has received 100,000 universal visitors. [August 2023]
- exoRBase has been updated to verson 2.0. [August 2021] [Version 1.0]
- exoRBase 2.0 is published in Database Issue of Nucleic Acids Research. [January 2022] [Full Text]
- exoRBase has received more than 70,000 universal visitors. [June 2021]
- EV-origin: enumerating the tissue-cellular origin of circulating extracellular vesicles using exLR profile, published in Comput Struct Biotechnol J. [October 2020][Full Text]
- The exLR biomarkers used for the detection of pancreatic ductal adenocarcinoma published in Gut. [March 2020] [Full Text]
- The feature and application of human blood exLRs published in Clin Chem. [March 2019] [Full Text]
- exoRBase has received 10,000 visitors. [May 2018]
- exoRBase is published in Database Issue of Nucleic Acids Research. [October 2017] [Full Text]
- exoRBase is publicly accessible. [August 2017]
- 77 experimental validations from 18 published literatures have been included. [July 2017]
- GTEx and circBase data have been used to annotate possible original cells/tissues of exosomal RNAs. [July 2017]
- 92 RNA-seq samples from seven experiments have been integrated into exoRBase. [June 2017]